Hinweis
Gehen Sie zum Ende, um den vollständigen Beispielcode herunterzuladen oder dieses Beispiel über JupyterLite oder Binder in Ihrem Browser auszuführen.
Wikipedia Haupteigenvektor#
Eine klassische Methode, um die relative Wichtigkeit von Knoten in einem Graphen zu ermitteln, ist die Berechnung des Haupteigenvektors der Adjazenzmatrix, um jedem Knoten die Werte der Komponenten des ersten Eigenvektors als Zentralitätswert zuzuweisen: https://en.wikipedia.org/wiki/Eigenvector_centrality. Bei dem Graphen von Webseiten und Links werden diese Werte von Google als PageRank-Scores bezeichnet.
Ziel dieses Beispiels ist es, den Linkgraphen innerhalb von Wikipedia-Artikeln zu analysieren, um Artikel nach relativer Wichtigkeit gemäß dieser Eigenvektor-Zentralität zu ordnen.
Die traditionelle Methode zur Berechnung des Haupteigenvektors ist die Potenziteration-Methode. Hier wird die Berechnung dank Martinsons Randomisiertem SVD-Algorithmus, implementiert in scikit-learn, erreicht.
Die Graphdaten werden aus den DBpedia-Dumps abgerufen. DBpedia ist eine Extraktion der latenten strukturierten Daten des Wikipedia-Inhalts.
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
import os
from bz2 import BZ2File
from datetime import datetime
from pprint import pprint
from time import time
from urllib.request import urlopen
import numpy as np
from scipy import sparse
from sklearn.decomposition import randomized_svd
Daten herunterladen, falls nicht bereits auf der Festplatte vorhanden#
redirects_url = "http://downloads.dbpedia.org/3.5.1/en/redirects_en.nt.bz2"
redirects_filename = redirects_url.rsplit("/", 1)[1]
page_links_url = "http://downloads.dbpedia.org/3.5.1/en/page_links_en.nt.bz2"
page_links_filename = page_links_url.rsplit("/", 1)[1]
resources = [
(redirects_url, redirects_filename),
(page_links_url, page_links_filename),
]
for url, filename in resources:
if not os.path.exists(filename):
print("Downloading data from '%s', please wait..." % url)
opener = urlopen(url)
with open(filename, "wb") as f:
f.write(opener.read())
print()
Laden der Weiterleitungsdateien#
def index(redirects, index_map, k):
"""Find the index of an article name after redirect resolution"""
k = redirects.get(k, k)
return index_map.setdefault(k, len(index_map))
DBPEDIA_RESOURCE_PREFIX_LEN = len("http://dbpedia.org/resource/")
SHORTNAME_SLICE = slice(DBPEDIA_RESOURCE_PREFIX_LEN + 1, -1)
def short_name(nt_uri):
"""Remove the < and > URI markers and the common URI prefix"""
return nt_uri[SHORTNAME_SLICE]
def get_redirects(redirects_filename):
"""Parse the redirections and build a transitively closed map out of it"""
redirects = {}
print("Parsing the NT redirect file")
for l, line in enumerate(BZ2File(redirects_filename)):
split = line.split()
if len(split) != 4:
print("ignoring malformed line: " + line)
continue
redirects[short_name(split[0])] = short_name(split[2])
if l % 1000000 == 0:
print("[%s] line: %08d" % (datetime.now().isoformat(), l))
# compute the transitive closure
print("Computing the transitive closure of the redirect relation")
for l, source in enumerate(redirects.keys()):
transitive_target = None
target = redirects[source]
seen = {source}
while True:
transitive_target = target
target = redirects.get(target)
if target is None or target in seen:
break
seen.add(target)
redirects[source] = transitive_target
if l % 1000000 == 0:
print("[%s] line: %08d" % (datetime.now().isoformat(), l))
return redirects
Berechnung der Adjazenzmatrix#
def get_adjacency_matrix(redirects_filename, page_links_filename, limit=None):
"""Extract the adjacency graph as a scipy sparse matrix
Redirects are resolved first.
Returns X, the scipy sparse adjacency matrix, redirects as python
dict from article names to article names and index_map a python dict
from article names to python int (article indexes).
"""
print("Computing the redirect map")
redirects = get_redirects(redirects_filename)
print("Computing the integer index map")
index_map = dict()
links = list()
for l, line in enumerate(BZ2File(page_links_filename)):
split = line.split()
if len(split) != 4:
print("ignoring malformed line: " + line)
continue
i = index(redirects, index_map, short_name(split[0]))
j = index(redirects, index_map, short_name(split[2]))
links.append((i, j))
if l % 1000000 == 0:
print("[%s] line: %08d" % (datetime.now().isoformat(), l))
if limit is not None and l >= limit - 1:
break
print("Computing the adjacency matrix")
X = sparse.lil_matrix((len(index_map), len(index_map)), dtype=np.float32)
for i, j in links:
X[i, j] = 1.0
del links
print("Converting to CSR representation")
X = X.tocsr()
print("CSR conversion done")
return X, redirects, index_map
# stop after 5M links to make it possible to work in RAM
X, redirects, index_map = get_adjacency_matrix(
redirects_filename, page_links_filename, limit=5000000
)
names = {i: name for name, i in index_map.items()}
Berechnung des Hauptsingulären Vektors mit Randomisiertem SVD#
print("Computing the principal singular vectors using randomized_svd")
t0 = time()
U, s, V = randomized_svd(X, 5, n_iter=3)
print("done in %0.3fs" % (time() - t0))
# print the names of the wikipedia related strongest components of the
# principal singular vector which should be similar to the highest eigenvector
print("Top wikipedia pages according to principal singular vectors")
pprint([names[i] for i in np.abs(U.T[0]).argsort()[-10:]])
pprint([names[i] for i in np.abs(V[0]).argsort()[-10:]])
Berechnung der Zentralitätswerte#
def centrality_scores(X, alpha=0.85, max_iter=100, tol=1e-10):
"""Power iteration computation of the principal eigenvector
This method is also known as Google PageRank and the implementation
is based on the one from the NetworkX project (BSD licensed too)
with copyrights by:
Aric Hagberg <hagberg@lanl.gov>
Dan Schult <dschult@colgate.edu>
Pieter Swart <swart@lanl.gov>
"""
n = X.shape[0]
X = X.copy()
incoming_counts = np.asarray(X.sum(axis=1)).ravel()
print("Normalizing the graph")
for i in incoming_counts.nonzero()[0]:
X.data[X.indptr[i] : X.indptr[i + 1]] *= 1.0 / incoming_counts[i]
dangle = np.asarray(np.where(np.isclose(X.sum(axis=1), 0), 1.0 / n, 0)).ravel()
scores = np.full(n, 1.0 / n, dtype=np.float32) # initial guess
for i in range(max_iter):
print("power iteration #%d" % i)
prev_scores = scores
scores = (
alpha * (scores * X + np.dot(dangle, prev_scores))
+ (1 - alpha) * prev_scores.sum() / n
)
# check convergence: normalized l_inf norm
scores_max = np.abs(scores).max()
if scores_max == 0.0:
scores_max = 1.0
err = np.abs(scores - prev_scores).max() / scores_max
print("error: %0.6f" % err)
if err < n * tol:
return scores
return scores
print("Computing principal eigenvector score using a power iteration method")
t0 = time()
scores = centrality_scores(X, max_iter=100)
print("done in %0.3fs" % (time() - t0))
pprint([names[i] for i in np.abs(scores).argsort()[-10:]])
Verwandte Beispiele
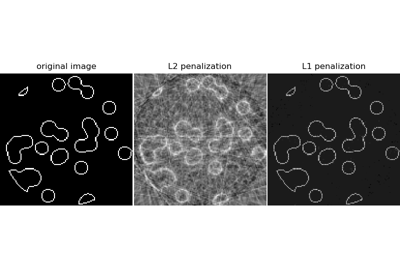
Kompression Sensing: Tomographie-Rekonstruktion mit L1-Prior (Lasso)
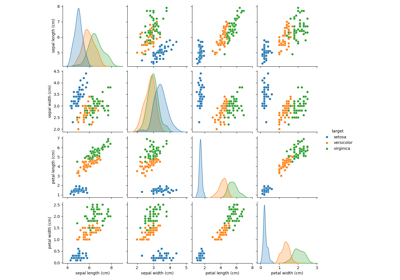
Principal Component Analysis (PCA) auf dem Iris-Datensatz

Segmentierung des Bildes von griechischen Münzen in Regionen